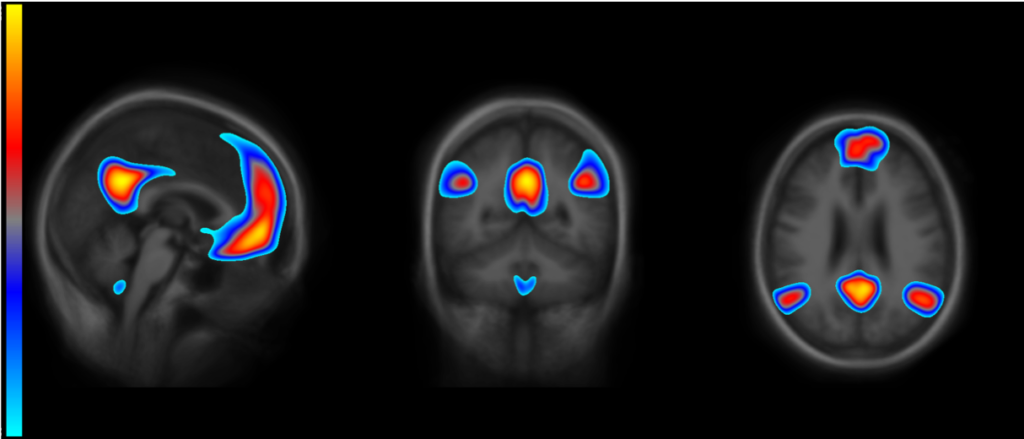
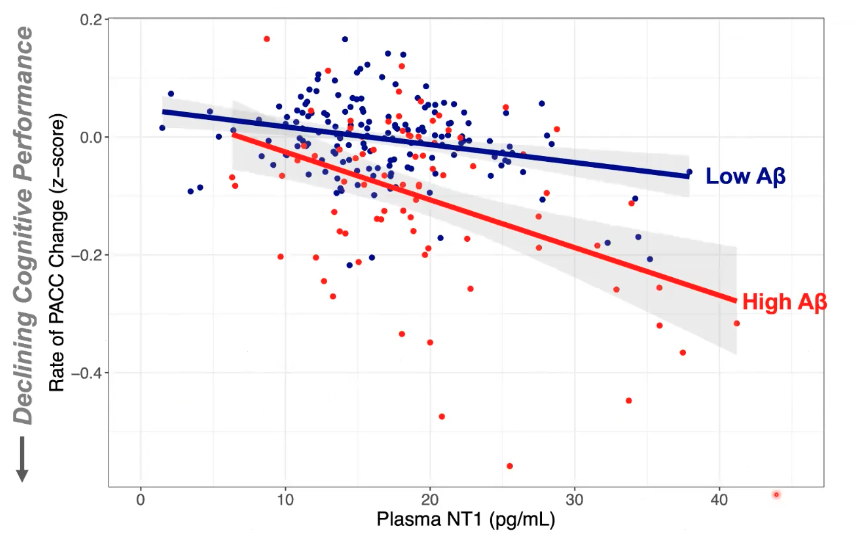
For Participants
Information for participants and those interested in participating
The Harvard Aging Brain Study (HABS) is an observational study that aims to find out whether the changes that a doctor sometimes sees on a brain scan are related to early memory changes that occur in older healthy adults.