This examples computes the coherence between a seed in the left auditory cortex and the rest of the brain based on single-trial MNE-dSPM inverse solutions.
Out:
Reading inverse operator decomposition from /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif...
Reading inverse operator info...
[done]
Reading inverse operator decomposition...
[done]
305 x 305 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Noise covariance matrix read.
22494 x 22494 diagonal covariance (kind = 2) found.
Source covariance matrix read.
22494 x 22494 diagonal covariance (kind = 6) found.
Orientation priors read.
22494 x 22494 diagonal covariance (kind = 5) found.
Depth priors read.
Did not find the desired covariance matrix (kind = 3)
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Source spaces transformed to the inverse solution coordinate frame
Opening raw data file /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Current compensation grade : 0
72 matching events found
Created an SSP operator (subspace dimension = 3)
4 projection items activated
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on MAG : [u'MEG 1711']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 55
Created the regularized inverter
Created an SSP operator (subspace dimension = 3)
Created the whitener using a full noise covariance matrix (3 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Picked 305 channels from the data
Computing inverse...
(eigenleads need to be weighted)...
(dSPM)...
[done]
Connectivity computation...
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 3)
Created the whitener using a full noise covariance matrix (3 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Picked 305 channels from the data
Computing inverse...
(eigenleads need to be weighted)...
Processing epoch : 1
computing connectivity for 7498 connections
using t=-0.200s..0.499s for estimation (106 points)
computing connectivity for the bands:
band 1: 8.5Hz..12.7Hz (4 points)
band 2: 14.2Hz..29.7Hz (12 points)
connectivity scores will be averaged for each band
using FFT with a Hanning window to estimate spectra
the following metrics will be computed: Coherence
computing connectivity for epoch 1
Processing epoch : 2
computing connectivity for epoch 2
Processing epoch : 3
computing connectivity for epoch 3
Processing epoch : 4
computing connectivity for epoch 4
Processing epoch : 5
computing connectivity for epoch 5
Processing epoch : 6
computing connectivity for epoch 6
Processing epoch : 7
computing connectivity for epoch 7
Processing epoch : 8
computing connectivity for epoch 8
Processing epoch : 9
computing connectivity for epoch 9
Processing epoch : 10
computing connectivity for epoch 10
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 11
computing connectivity for epoch 11
Processing epoch : 12
computing connectivity for epoch 12
Processing epoch : 13
computing connectivity for epoch 13
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 14
computing connectivity for epoch 14
Processing epoch : 15
computing connectivity for epoch 15
Processing epoch : 16
computing connectivity for epoch 16
Processing epoch : 17
computing connectivity for epoch 17
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 18
computing connectivity for epoch 18
Processing epoch : 19
computing connectivity for epoch 19
Processing epoch : 20
computing connectivity for epoch 20
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 21
computing connectivity for epoch 21
Processing epoch : 22
computing connectivity for epoch 22
Processing epoch : 23
computing connectivity for epoch 23
Rejecting epoch based on MAG : [u'MEG 1711']
Processing epoch : 24
computing connectivity for epoch 24
Processing epoch : 25
computing connectivity for epoch 25
Processing epoch : 26
computing connectivity for epoch 26
Processing epoch : 27
computing connectivity for epoch 27
Processing epoch : 28
computing connectivity for epoch 28
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 29
computing connectivity for epoch 29
Processing epoch : 30
computing connectivity for epoch 30
Processing epoch : 31
computing connectivity for epoch 31
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 32
computing connectivity for epoch 32
Processing epoch : 33
computing connectivity for epoch 33
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 34
computing connectivity for epoch 34
Processing epoch : 35
computing connectivity for epoch 35
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 36
computing connectivity for epoch 36
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 37
computing connectivity for epoch 37
Processing epoch : 38
computing connectivity for epoch 38
Processing epoch : 39
computing connectivity for epoch 39
Processing epoch : 40
computing connectivity for epoch 40
Processing epoch : 41
computing connectivity for epoch 41
Processing epoch : 42
computing connectivity for epoch 42
Processing epoch : 43
computing connectivity for epoch 43
Processing epoch : 44
computing connectivity for epoch 44
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 45
computing connectivity for epoch 45
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 46
computing connectivity for epoch 46
Processing epoch : 47
computing connectivity for epoch 47
Processing epoch : 48
computing connectivity for epoch 48
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 49
computing connectivity for epoch 49
Processing epoch : 50
computing connectivity for epoch 50
Processing epoch : 51
computing connectivity for epoch 51
Processing epoch : 52
computing connectivity for epoch 52
Processing epoch : 53
computing connectivity for epoch 53
Processing epoch : 54
computing connectivity for epoch 54
Processing epoch : 55
computing connectivity for epoch 55
[done]
[Connectivity computation done]
Frequencies in Hz over which coherence was averaged for alpha:
[ 8.49926873 9.91581352 11.33235831 12.74890309]
Frequencies in Hz over which coherence was averaged for beta:
[ 14.16544788 15.58199267 16.99853746 18.41508225 19.83162704
21.24817182 22.66471661 24.0812614 25.49780619 26.91435098
28.33089577 29.74744055]
Updating smoothing matrix, be patient..
Smoothing matrix creation, step 1
Smoothing matrix creation, step 2
Smoothing matrix creation, step 3
Smoothing matrix creation, step 4
Smoothing matrix creation, step 5
Smoothing matrix creation, step 6
Smoothing matrix creation, step 7
Smoothing matrix creation, step 8
Smoothing matrix creation, step 9
Smoothing matrix creation, step 10
colormap: fmin=2.50e-01 fmid=4.00e-01 fmax=6.50e-01 transparent=1
Updating smoothing matrix, be patient..
Smoothing matrix creation, step 1
Smoothing matrix creation, step 2
Smoothing matrix creation, step 3
Smoothing matrix creation, step 4
Smoothing matrix creation, step 5
Smoothing matrix creation, step 6
Smoothing matrix creation, step 7
Smoothing matrix creation, step 8
Smoothing matrix creation, step 9
Smoothing matrix creation, step 10
colormap: fmin=2.50e-01 fmid=4.00e-01 fmax=6.50e-01 transparent=1
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
import numpy as np
import mne
from mne.datasets import sample
from mne.minimum_norm import (apply_inverse, apply_inverse_epochs,
read_inverse_operator)
from mne.connectivity import seed_target_indices, spectral_connectivity
print(__doc__)
data_path = sample.data_path()
subjects_dir = data_path + '/subjects'
fname_inv = data_path + '/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif'
fname_raw = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
fname_event = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
label_name_lh = 'Aud-lh'
fname_label_lh = data_path + '/MEG/sample/labels/%s.label' % label_name_lh
event_id, tmin, tmax = 1, -0.2, 0.5
method = "dSPM" # use dSPM method (could also be MNE or sLORETA)
# Load data.
inverse_operator = read_inverse_operator(fname_inv)
label_lh = mne.read_label(fname_label_lh)
raw = mne.io.read_raw_fif(fname_raw)
events = mne.read_events(fname_event)
# Add a bad channel.
raw.info['bads'] += ['MEG 2443']
# pick MEG channels.
picks = mne.pick_types(raw.info, meg=True, eeg=False, stim=False, eog=True,
exclude='bads')
# Read epochs.
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, picks=picks,
baseline=(None, 0),
reject=dict(mag=4e-12, grad=4000e-13, eog=150e-6))
# First, we find the most active vertex in the left auditory cortex, which
# we will later use as seed for the connectivity computation.
snr = 3.0
lambda2 = 1.0 / snr ** 2
evoked = epochs.average()
stc = apply_inverse(evoked, inverse_operator, lambda2, method,
pick_ori="normal")
# Restrict the source estimate to the label in the left auditory cortex.
stc_label = stc.in_label(label_lh)
# Find number and index of vertex with most power.
src_pow = np.sum(stc_label.data ** 2, axis=1)
seed_vertno = stc_label.vertices[0][np.argmax(src_pow)]
seed_idx = np.searchsorted(stc.vertices[0], seed_vertno) # index in orig stc
# Generate index parameter for seed-based connectivity analysis.
n_sources = stc.data.shape[0]
indices = seed_target_indices([seed_idx], np.arange(n_sources))
# Compute inverse solution and for each epoch. By using "return_generator=True"
# stcs will be a generator object instead of a list. This allows us so to
# compute the coherence without having to keep all source estimates in memory.
snr = 1.0 # use lower SNR for single epochs
lambda2 = 1.0 / snr ** 2
stcs = apply_inverse_epochs(epochs, inverse_operator, lambda2, method,
pick_ori="normal", return_generator=True)
# Now we are ready to compute the coherence in the alpha and beta band.
# fmin and fmax specify the lower and upper freq. for each band, resp.
fmin = (8., 13.)
fmax = (13., 30.)
sfreq = raw.info['sfreq'] # the sampling frequency
# Now we compute connectivity. To speed things up, we use 2 parallel jobs
# and use mode='fourier', which uses a FFT with a Hanning window
# to compute the spectra (instead of multitaper estimation, which has a
# lower variance but is slower). By using faverage=True, we directly
# average the coherence in the alpha and beta band, i.e., we will only
# get 2 frequency bins.
coh, freqs, times, n_epochs, n_tapers = spectral_connectivity(
stcs, method='coh', mode='fourier', indices=indices,
sfreq=sfreq, fmin=fmin, fmax=fmax, faverage=True, n_jobs=1)
print('Frequencies in Hz over which coherence was averaged for alpha: ')
print(freqs[0])
print('Frequencies in Hz over which coherence was averaged for beta: ')
print(freqs[1])
# Generate a SourceEstimate with the coherence. This is simple since we
# used a single seed. For more than one seeds we would have to split coh.
# Note: We use a hack to save the frequency axis as time.
tmin = np.mean(freqs[0])
tstep = np.mean(freqs[1]) - tmin
coh_stc = mne.SourceEstimate(coh, vertices=stc.vertices, tmin=1e-3 * tmin,
tstep=1e-3 * tstep, subject='sample')
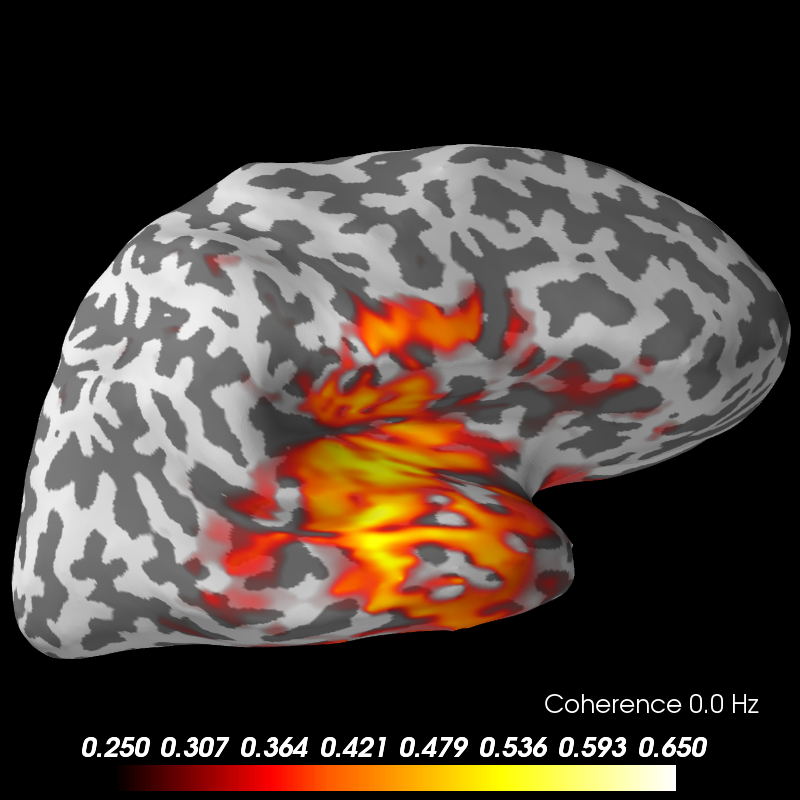
# Now we can visualize the coherence using the plot method.
brain = coh_stc.plot('sample', 'inflated', 'both',
time_label='Coherence %0.1f Hz',
subjects_dir=subjects_dir,
clim=dict(kind='value', lims=(0.25, 0.4, 0.65)))
brain.show_view('lateral')
Total running time of the script: ( 0 minutes 40.514 seconds)