Compute dSPM inverse solution on single trial epochs restricted to a brain label.
# Author: Alexandre Gramfort <alexandre.gramfort@telecom-paristech.fr>
#
# License: BSD (3-clause)
import numpy as np
import matplotlib.pyplot as plt
import mne
from mne.datasets import sample
from mne.minimum_norm import apply_inverse_epochs, read_inverse_operator
from mne.minimum_norm import apply_inverse
print(__doc__)
data_path = sample.data_path()
fname_inv = data_path + '/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif'
fname_raw = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
fname_event = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
label_name = 'Aud-lh'
fname_label = data_path + '/MEG/sample/labels/%s.label' % label_name
event_id, tmin, tmax = 1, -0.2, 0.5
# Using the same inverse operator when inspecting single trials Vs. evoked
snr = 3.0 # Standard assumption for average data but using it for single trial
lambda2 = 1.0 / snr ** 2
method = "dSPM" # use dSPM method (could also be MNE or sLORETA)
# Load data
inverse_operator = read_inverse_operator(fname_inv)
label = mne.read_label(fname_label)
raw = mne.io.read_raw_fif(fname_raw)
events = mne.read_events(fname_event)
# Set up pick list
include = []
# Add a bad channel
raw.info['bads'] += ['EEG 053'] # bads + 1 more
# pick MEG channels
picks = mne.pick_types(raw.info, meg=True, eeg=False, stim=False, eog=True,
include=include, exclude='bads')
# Read epochs
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, picks=picks,
baseline=(None, 0), reject=dict(mag=4e-12, grad=4000e-13,
eog=150e-6))
# Get evoked data (averaging across trials in sensor space)
evoked = epochs.average()
# Compute inverse solution and stcs for each epoch
# Use the same inverse operator as with evoked data (i.e., set nave)
# If you use a different nave, dSPM just scales by a factor sqrt(nave)
stcs = apply_inverse_epochs(epochs, inverse_operator, lambda2, method, label,
pick_ori="normal", nave=evoked.nave)
stc_evoked = apply_inverse(evoked, inverse_operator, lambda2, method,
pick_ori="normal")
stc_evoked_label = stc_evoked.in_label(label)
# Mean across trials but not across vertices in label
mean_stc = sum(stcs) / len(stcs)
# compute sign flip to avoid signal cancellation when averaging signed values
flip = mne.label_sign_flip(label, inverse_operator['src'])
label_mean = np.mean(mean_stc.data, axis=0)
label_mean_flip = np.mean(flip[:, np.newaxis] * mean_stc.data, axis=0)
# Get inverse solution by inverting evoked data
stc_evoked = apply_inverse(evoked, inverse_operator, lambda2, method,
pick_ori="normal")
# apply_inverse() does whole brain, so sub-select label of interest
stc_evoked_label = stc_evoked.in_label(label)
# Average over label (not caring to align polarities here)
label_mean_evoked = np.mean(stc_evoked_label.data, axis=0)
Out:
Reading inverse operator decomposition from /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif...
Reading inverse operator info...
[done]
Reading inverse operator decomposition...
[done]
305 x 305 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Noise covariance matrix read.
22494 x 22494 diagonal covariance (kind = 2) found.
Source covariance matrix read.
22494 x 22494 diagonal covariance (kind = 6) found.
Orientation priors read.
22494 x 22494 diagonal covariance (kind = 5) found.
Depth priors read.
Did not find the desired covariance matrix (kind = 3)
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Source spaces transformed to the inverse solution coordinate frame
Opening raw data file /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Current compensation grade : 0
72 matching events found
Created an SSP operator (subspace dimension = 3)
4 projection items activated
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on MAG : [u'MEG 1711']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 55
Created the regularized inverter
Created an SSP operator (subspace dimension = 3)
Created the whitener using a full noise covariance matrix (3 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Picked 305 channels from the data
Computing inverse...
(eigenleads need to be weighted)...
Processing epoch : 1
Processing epoch : 2
Processing epoch : 3
Processing epoch : 4
Processing epoch : 5
Processing epoch : 6
Processing epoch : 7
Processing epoch : 8
Processing epoch : 9
Processing epoch : 10
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 11
Processing epoch : 12
Processing epoch : 13
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 14
Processing epoch : 15
Processing epoch : 16
Processing epoch : 17
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 18
Processing epoch : 19
Processing epoch : 20
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 21
Processing epoch : 22
Processing epoch : 23
Rejecting epoch based on MAG : [u'MEG 1711']
Processing epoch : 24
Processing epoch : 25
Processing epoch : 26
Processing epoch : 27
Processing epoch : 28
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 29
Processing epoch : 30
Processing epoch : 31
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 32
Processing epoch : 33
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 34
Processing epoch : 35
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 36
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 37
Processing epoch : 38
Processing epoch : 39
Processing epoch : 40
Processing epoch : 41
Processing epoch : 42
Processing epoch : 43
Processing epoch : 44
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 45
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 46
Processing epoch : 47
Processing epoch : 48
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on EOG : [u'EOG 061']
Processing epoch : 49
Processing epoch : 50
Processing epoch : 51
Processing epoch : 52
Processing epoch : 53
Processing epoch : 54
Processing epoch : 55
[done]
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 55
Created the regularized inverter
Created an SSP operator (subspace dimension = 3)
Created the whitener using a full noise covariance matrix (3 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Picked 305 channels from the data
Computing inverse...
(eigenleads need to be weighted)...
(dSPM)...
[done]
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 55
Created the regularized inverter
Created an SSP operator (subspace dimension = 3)
Created the whitener using a full noise covariance matrix (3 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Picked 305 channels from the data
Computing inverse...
(eigenleads need to be weighted)...
(dSPM)...
[done]
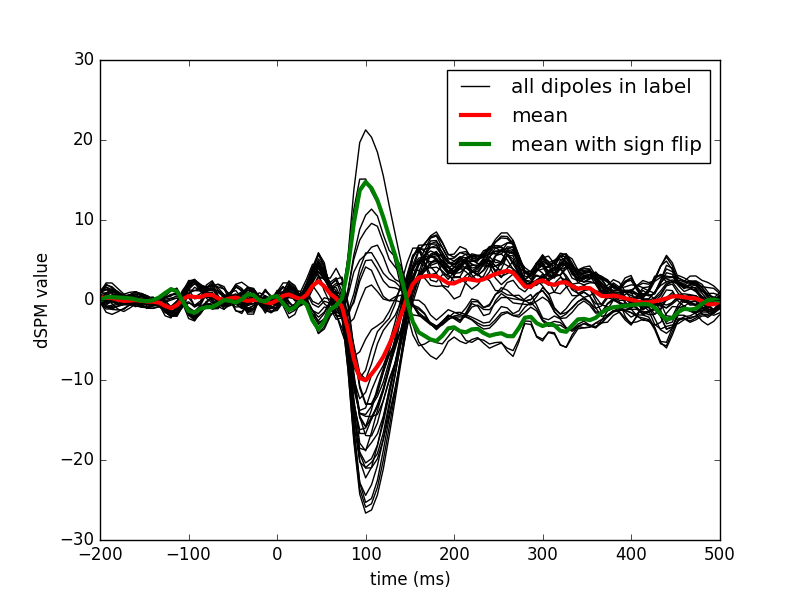
View activation time-series to illustrate the benefit of aligning/flipping
times = 1e3 * stcs[0].times # times in ms
plt.figure()
h0 = plt.plot(times, mean_stc.data.T, 'k')
h1, = plt.plot(times, label_mean, 'r', linewidth=3)
h2, = plt.plot(times, label_mean_flip, 'g', linewidth=3)
plt.legend((h0[0], h1, h2), ('all dipoles in label', 'mean',
'mean with sign flip'))
plt.xlabel('time (ms)')
plt.ylabel('dSPM value')
plt.show()
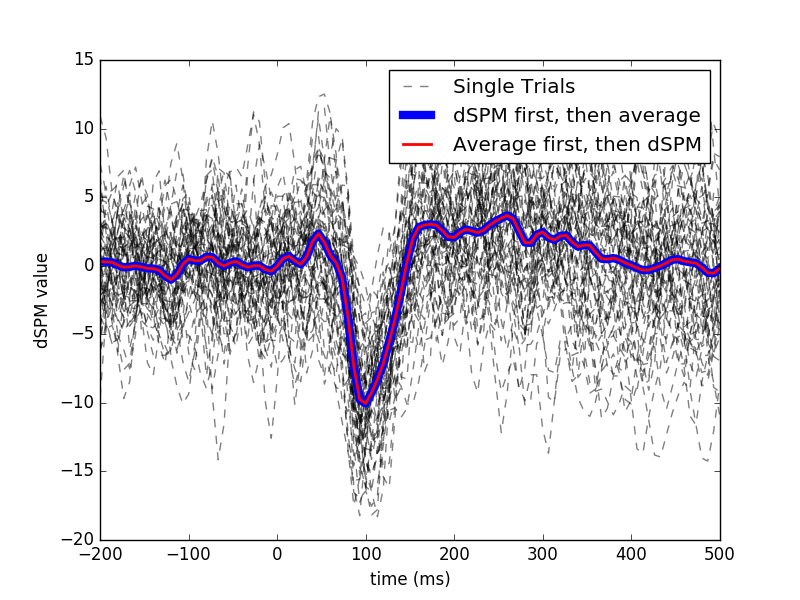
Viewing single trial dSPM and average dSPM for unflipped pooling over label Compare to (1) Inverse (dSPM) then average, (2) Evoked then dSPM
# Single trial
plt.figure()
for k, stc_trial in enumerate(stcs):
plt.plot(times, np.mean(stc_trial.data, axis=0).T, 'k--',
label='Single Trials' if k == 0 else '_nolegend_',
alpha=0.5)
# Single trial inverse then average.. making linewidth large to not be masked
plt.plot(times, label_mean, 'b', linewidth=6,
label='dSPM first, then average')
# Evoked and then inverse
plt.plot(times, label_mean_evoked, 'r', linewidth=2,
label='Average first, then dSPM')
plt.xlabel('time (ms)')
plt.ylabel('dSPM value')
plt.legend()
plt.show()
Total running time of the script: ( 0 minutes 2.846 seconds)