Locate QRS component of ECG.
# Authors: Alexandre Gramfort <alexandre.gramfort@telecom-paristech.fr>
#
# License: BSD (3-clause)
import numpy as np
import matplotlib.pyplot as plt
import mne
from mne import io
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
Set parameters
raw_fname = data_path + '/MEG/sample/sample_audvis_raw.fif'
# Setup for reading the raw data
raw = io.read_raw_fif(raw_fname)
event_id = 999
ecg_events, _, _ = mne.preprocessing.find_ecg_events(raw, event_id,
ch_name='MEG 1531')
# Read epochs
picks = mne.pick_types(raw.info, meg=False, eeg=False, stim=False, eog=False,
include=['MEG 1531'], exclude='bads')
tmin, tmax = -0.1, 0.1
epochs = mne.Epochs(raw, ecg_events, event_id, tmin, tmax, picks=picks,
proj=False)
data = epochs.get_data()
print("Number of detected ECG artifacts : %d" % len(data))
Out:
Opening raw data file /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Current compensation grade : 0
Using channel MEG 1531 to identify heart beats.
Setting up band-pass filter from 5 - 35 Hz
Number of ECG events detected : 284 (average pulse 61 / min.)
284 matching events found
Projection vector "PCA-v1" has magnitude 0.08 (should be unity), applying projector with 1/102 of the original channels available may be dangerous, consider recomputing and adding projection vectors for channels that are eventually used. If this is intentional, consider using info.normalize_proj()
Projection vector "PCA-v2" has magnitude 0.00 (should be unity), applying projector with 1/102 of the original channels available may be dangerous, consider recomputing and adding projection vectors for channels that are eventually used. If this is intentional, consider using info.normalize_proj()
Projection vector "PCA-v3" has magnitude 0.13 (should be unity), applying projector with 1/102 of the original channels available may be dangerous, consider recomputing and adding projection vectors for channels that are eventually used. If this is intentional, consider using info.normalize_proj()
Created an SSP operator (subspace dimension = 1)
Loading data for 284 events and 121 original time points ...
0 bad epochs dropped
Number of detected ECG artifacts : 284
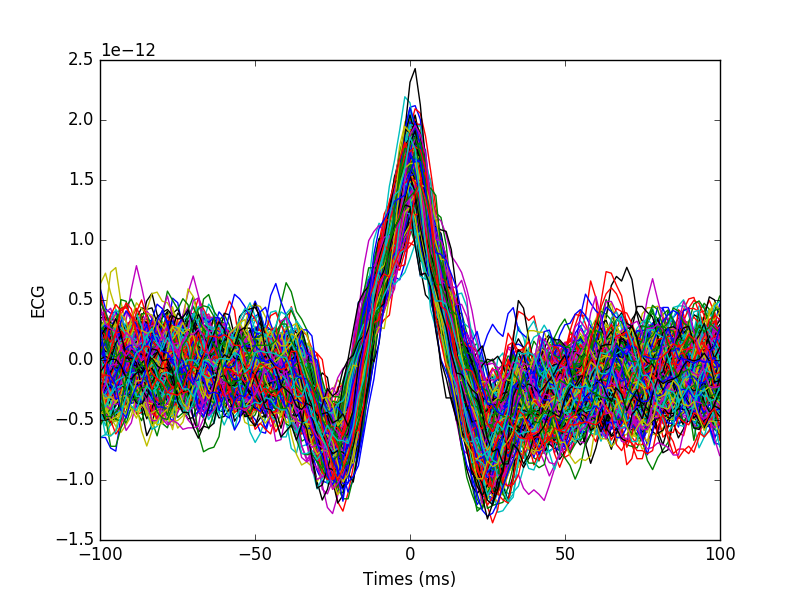
Plot ECG artifacts
plt.plot(1e3 * epochs.times, np.squeeze(data).T)
plt.xlabel('Times (ms)')
plt.ylabel('ECG')
plt.show()
Total running time of the script: ( 0 minutes 2.018 seconds)