When performing experiments where timing is critical, a signal with a high sampling rate is desired. However, having a signal with a much higher sampling rate than is necessary needlessly consumes memory and slows down computations operating on the data.
This example downsamples from 600 Hz to 100 Hz. This achieves a 6-fold reduction in data size, at the cost of an equal loss of temporal resolution.
# Authors: Marijn van Vliet <w.m.vanvliet@gmail.com>
#
# License: BSD (3-clause)
#
from __future__ import print_function
from matplotlib import pyplot as plt
import mne
from mne.datasets import sample
Setting up data paths and loading raw data (skip some data for speed)
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_raw.fif'
raw = mne.io.read_raw_fif(raw_fname).crop(120, 240).load_data()
Out:
Opening raw data file /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Current compensation grade : 0
Reading 0 ... 72074 = 0.000 ... 120.000 secs...
Since downsampling reduces the timing precision of events, we recommend first extracting epochs and downsampling the Epochs object:
events = mne.find_events(raw)
epochs = mne.Epochs(raw, events, event_id=2, tmin=-0.1, tmax=0.8, preload=True)
# Downsample to 100 Hz
print('Original sampling rate:', epochs.info['sfreq'], 'Hz')
epochs_resampled = epochs.copy().resample(100, npad='auto')
print('New sampling rate:', epochs_resampled.info['sfreq'], 'Hz')
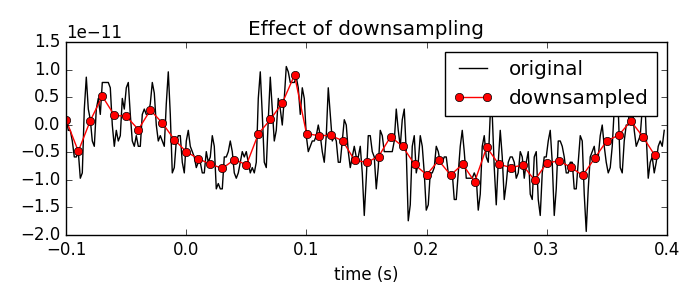
# Plot a piece of data to see the effects of downsampling
plt.figure(figsize=(7, 3))
n_samples_to_plot = int(0.5 * epochs.info['sfreq']) # plot 0.5 seconds of data
plt.plot(epochs.times[:n_samples_to_plot],
epochs.get_data()[0, 0, :n_samples_to_plot], color='black')
n_samples_to_plot = int(0.5 * epochs_resampled.info['sfreq'])
plt.plot(epochs_resampled.times[:n_samples_to_plot],
epochs_resampled.get_data()[0, 0, :n_samples_to_plot],
'-o', color='red')
plt.xlabel('time (s)')
plt.legend(['original', 'downsampled'], loc='best')
plt.title('Effect of downsampling')
mne.viz.tight_layout()
Out:
144 events found
Events id: [ 1 2 3 4 5 32]
33 matching events found
Created an SSP operator (subspace dimension = 3)
3 projection items activated
Loading data for 33 events and 541 original time points ...
0 bad epochs dropped
('Original sampling rate:', 600.614990234375, 'Hz')
('New sampling rate:', 100.0, 'Hz')
When resampling epochs is unwanted or impossible, for example when the data doesn’t fit into memory or your analysis pipeline doesn’t involve epochs at all, the alternative approach is to resample the continuous data. This can also be done on non-preloaded data.
# Resample to 300 Hz
raw_resampled = raw.copy().resample(300, npad='auto')
Out:
144 events found
Events id: [ 1 2 3 4 5 32]
144 events found
Events id: [ 1 2 3 4 5 32]
Because resampling also affects the stim channels, some trigger onsets might be lost in this case. While MNE attempts to downsample the stim channels in an intelligent manner to avoid this, the recommended approach is to find events on the original data before downsampling.
print('Number of events before resampling:', len(mne.find_events(raw)))
# Resample to 100 Hz (generates warning)
raw_resampled = raw.copy().resample(100, npad='auto')
print('Number of events after resampling:',
len(mne.find_events(raw_resampled)))
# To avoid losing events, jointly resample the data and event matrix
events = mne.find_events(raw)
raw_resampled, events_resampled = raw.copy().resample(
100, npad='auto', events=events)
print('Number of events after resampling:', len(events_resampled))
Out:
144 events found
Events id: [ 1 2 3 4 5 32]
('Number of events before resampling:', 144)
144 events found
Events id: [ 1 2 3 4 5 32]
143 events found
Events id: [ 1 2 3 4 5 32]
Resampling of the stim channels caused event information to become unreliable. Consider finding events on the original data and passing the event matrix as a parameter.
143 events found
Events id: [ 1 2 3 4 5 32]
('Number of events after resampling:', 143)
144 events found
Events id: [ 1 2 3 4 5 32]
('Number of events after resampling:', 144)
Total running time of the script: ( 0 minutes 15.634 seconds)