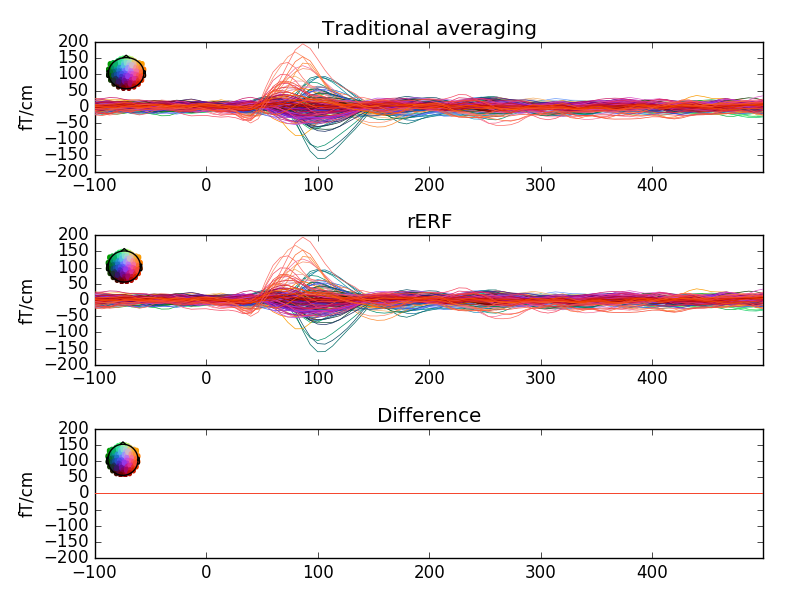
This demonstrates how rER[P/F]s - regressing the continuous data - is a generalisation of traditional averaging. If all preprocessing steps are the same, no overlap between epochs exists, and if all predictors are binary, regression is virtually identical to traditional averaging. If overlap exists and/or predictors are continuous, traditional averaging is inapplicable, but regression can estimate effects, including those of continuous predictors.
rERPs are described in: Smith, N. J., & Kutas, M. (2015). Regression-based estimation of ERP waveforms: II. Non-linear effects, overlap correction, and practical considerations. Psychophysiology, 52(2), 169-189.
Out:
Opening raw data file /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Current compensation grade : 0
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
Setting up high-pass filter at 1 Hz
l_trans_bandwidth chosen to be 1.0 Hz
Filter length of 991 samples (6.600 sec) selected
319 events found
Events id: [ 1 2 3 4 5 32]
# Authors: Jona Sassenhagen <jona.sassenhagen@gmail.de>
#
# License: BSD (3-clause)
import matplotlib.pyplot as plt
import mne
from mne.datasets import sample
from mne.stats.regression import linear_regression_raw
# Load and preprocess data
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw = mne.io.read_raw_fif(raw_fname, preload=True).pick_types(
meg='grad', stim=True, eeg=False).filter(1, None) # high-pass
# Set up events
events = mne.find_events(raw)
event_id = {'Aud/L': 1, 'Aud/R': 2}
tmin, tmax = -.1, .5
# regular epoching
picks = mne.pick_types(raw.info, meg=True)
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, reject=None,
baseline=None, preload=True, verbose=False)
# rERF
evokeds = linear_regression_raw(raw, events=events, event_id=event_id,
reject=None, tmin=tmin, tmax=tmax)
# linear_regression_raw returns a dict of evokeds
# select conditions similarly to mne.Epochs objects
# plot both results, and their difference
cond = "Aud/L"
fig, (ax1, ax2, ax3) = plt.subplots(3, 1)
params = dict(spatial_colors=True, show=False, ylim=dict(grad=(-200, 200)))
epochs[cond].average().plot(axes=ax1, **params)
evokeds[cond].plot(axes=ax2, **params)
contrast = mne.combine_evoked([evokeds[cond], -epochs[cond].average()],
weights='equal')
contrast.plot(axes=ax3, **params)
ax1.set_title("Traditional averaging")
ax2.set_title("rERF")
ax3.set_title("Difference")
plt.show()
Total running time of the script: ( 0 minutes 4.941 seconds)