Raw data structure: continuous data¶from __future__ import print_function
import mne
import os.path as op
from matplotlib import pyplot as plt
Continuous data is stored in objects of type Raw.
The core data structure is simply a 2D numpy array (channels × samples,
stored in a private attribute called ._data) combined with an
Info object (.info attribute)
(see The Info data structure).
The most common way to load continuous data is from a .fif file. For more information on loading data from other formats, or creating it from scratch.
# Load an example dataset, the preload flag loads the data into memory now
data_path = op.join(mne.datasets.sample.data_path(), 'MEG',
'sample', 'sample_audvis_raw.fif')
raw = mne.io.read_raw_fif(data_path, preload=True)
raw.set_eeg_reference() # set EEG average reference
# Give the sample rate
print('sample rate:', raw.info['sfreq'], 'Hz')
# Give the size of the data matrix
print('channels x samples:', raw._data.shape)
Out:
Opening raw data file /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Current compensation grade : 0
Reading 0 ... 166799 = 0.000 ... 277.714 secs...
Adding average EEG reference projection.
1 projection items deactivated
Average reference projection was added, but hasn't been applied yet. Use the .apply_proj() method function to apply projections.
('sample rate:', 600.614990234375, 'Hz')
('channels x samples:', (376, 166800))
Note
Accessing the ._data attribute is done here for educational purposes. However this is a private attribute as its name starts with an _. This suggests that you should not access this variable directly but rely on indexing syntax detailed just below.
Information about the channels contained in the Raw
object is contained in the Info attribute.
This is essentially a dictionary with a number of relevant fields (see
The Info data structure).
To access the data stored within Raw objects,
it is possible to index the Raw object.
Indexing a Raw object will return two arrays: an array
of times, as well as the data representing those timepoints. This works
even if the data is not preloaded, in which case the data will be read from
disk when indexing. The syntax is as follows:
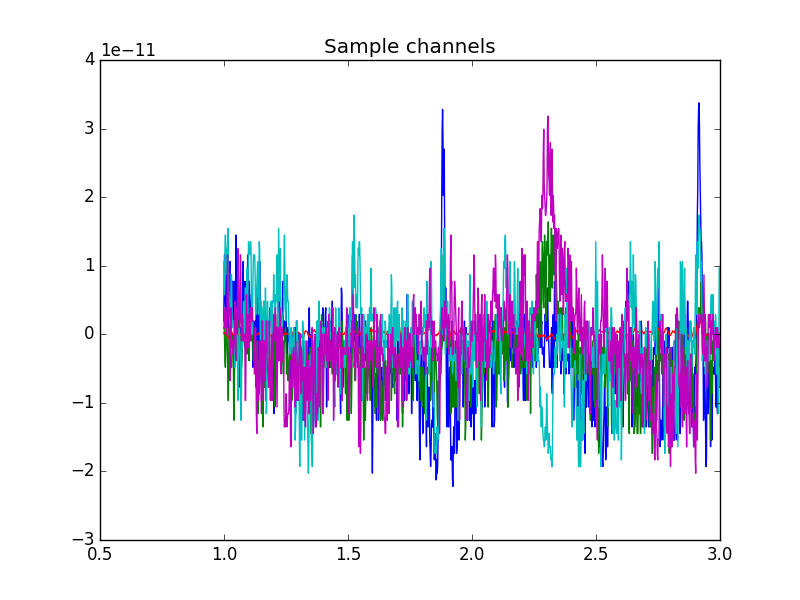
It is possible to use more intelligent indexing to extract data, using channel names, types or time ranges.
# Pull all MEG gradiometer channels:
# Make sure to use .copy() or it will overwrite the data
meg_only = raw.copy().pick_types(meg=True)
eeg_only = raw.copy().pick_types(meg=False, eeg=True)
# The MEG flag in particular lets you specify a string for more specificity
grad_only = raw.copy().pick_types(meg='grad')
# Or you can use custom channel names
pick_chans = ['MEG 0112', 'MEG 0111', 'MEG 0122', 'MEG 0123']
specific_chans = raw.copy().pick_channels(pick_chans)
print(meg_only)
print(eeg_only)
print(grad_only)
print(specific_chans)
Out:
<Raw | sample_audvis_raw.fif, n_channels x n_times : 305 x 166800 (277.7 sec), ~391.8 MB, data loaded>
<Raw | sample_audvis_raw.fif, n_channels x n_times : 59 x 166800 (277.7 sec), ~78.2 MB, data loaded>
<Raw | sample_audvis_raw.fif, n_channels x n_times : 203 x 166800 (277.7 sec), ~261.8 MB, data loaded>
<Raw | sample_audvis_raw.fif, n_channels x n_times : 4 x 166800 (277.7 sec), ~8.1 MB, data loaded>
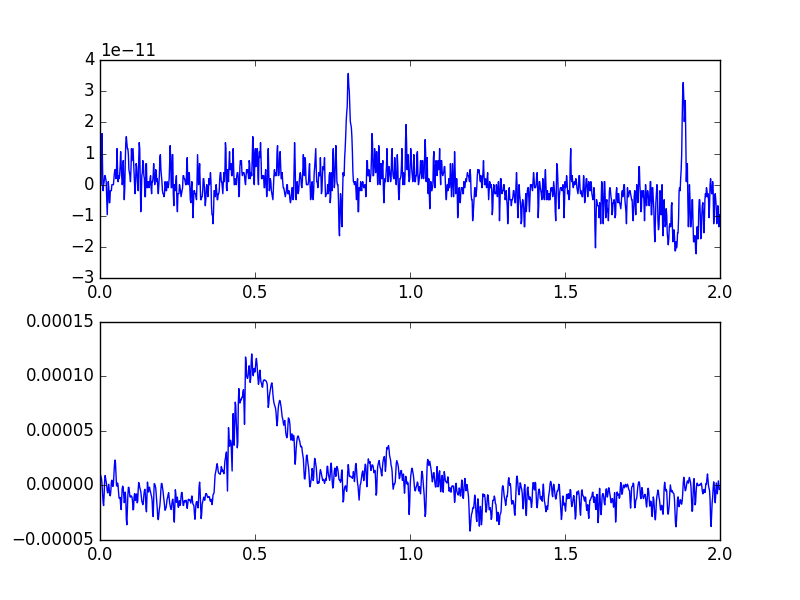
Notice the different scalings of these types
f, (a1, a2) = plt.subplots(2, 1)
eeg, times = eeg_only[0, :int(sfreq * 2)]
meg, times = meg_only[0, :int(sfreq * 2)]
a1.plot(times, meg[0])
a2.plot(times, eeg[0])
del eeg, meg, meg_only, grad_only, eeg_only, data, specific_chans
You can restrict the data to a specific time range
raw = raw.crop(0, 50) # in seconds
print('New time range from', raw.times.min(), 's to', raw.times.max(), 's')
Out:
('New time range from', 0.0, 's to', 50.00041705299622, 's')
And drop channels by name
nchan = raw.info['nchan']
raw = raw.drop_channels(['MEG 0241', 'EEG 001'])
print('Number of channels reduced from', nchan, 'to', raw.info['nchan'])
Out:
('Number of channels reduced from', 376, 'to', 374)
Raw objects¶Raw objects can be concatenated in time by using the
append function. For this to work, they must
have the same number of channels and their Info structures should be compatible.
# Create multiple :class:`Raw <mne.io.RawFIF>` objects
raw1 = raw.copy().crop(0, 10)
raw2 = raw.copy().crop(10, 20)
raw3 = raw.copy().crop(20, 40)
# Concatenate in time (also works without preloading)
raw1.append([raw2, raw3])
print('Time extends from', raw1.times.min(), 's to', raw1.times.max(), 's')
Out:
('Time extends from', 0.0, 's to', 40.003996554638213, 's')
Total running time of the script: ( 0 minutes 2.626 seconds)