Runs a full pipeline using MNE-Python:
- artifact removal
- averaging Epochs
- forward model computation
- source reconstruction using dSPM on the contrast : “faces - scrambled”
Note
This example does quite a bit of processing, so even on a fast machine it can take several minutes to complete.
# Authors: Alexandre Gramfort <alexandre.gramfort@telecom-paristech.fr>
# Denis Engemann <denis.engemann@gmail.com>
#
# License: BSD (3-clause)
import os.path as op
import matplotlib.pyplot as plt
import mne
from mne.datasets import spm_face
from mne.preprocessing import ICA, create_eog_epochs
from mne import io, combine_evoked
from mne.minimum_norm import make_inverse_operator, apply_inverse
print(__doc__)
data_path = spm_face.data_path()
subjects_dir = data_path + '/subjects'
Load and filter data, set up epochs
raw_fname = data_path + '/MEG/spm/SPM_CTF_MEG_example_faces%d_3D.ds'
raw = io.read_raw_ctf(raw_fname % 1, preload=True) # Take first run
# Here to save memory and time we'll downsample heavily -- this is not
# advised for real data as it can effectively jitter events!
raw.resample(120., npad='auto')
picks = mne.pick_types(raw.info, meg=True, exclude='bads')
raw.filter(1, 30, method='iir')
events = mne.find_events(raw, stim_channel='UPPT001')
# plot the events to get an idea of the paradigm
mne.viz.plot_events(events, raw.info['sfreq'])
event_ids = {"faces": 1, "scrambled": 2}
tmin, tmax = -0.2, 0.6
baseline = None # no baseline as high-pass is applied
reject = dict(mag=5e-12)
epochs = mne.Epochs(raw, events, event_ids, tmin, tmax, picks=picks,
baseline=baseline, preload=True, reject=reject)
# Fit ICA, find and remove major artifacts
ica = ICA(n_components=0.95, random_state=0).fit(raw, decim=1, reject=reject)
# compute correlation scores, get bad indices sorted by score
eog_epochs = create_eog_epochs(raw, ch_name='MRT31-2908', reject=reject)
eog_inds, eog_scores = ica.find_bads_eog(eog_epochs, ch_name='MRT31-2908')
ica.plot_scores(eog_scores, eog_inds) # see scores the selection is based on
ica.plot_components(eog_inds) # view topographic sensitivity of components
ica.exclude += eog_inds[:1] # we saw the 2nd ECG component looked too dipolar
ica.plot_overlay(eog_epochs.average()) # inspect artifact removal
ica.apply(epochs) # clean data, default in place
evoked = [epochs[k].average() for k in event_ids]
contrast = combine_evoked(evoked, weights=[-1, 1]) # Faces - scrambled
evoked.append(contrast)
for e in evoked:
e.plot(ylim=dict(mag=[-400, 400]))
plt.show()
# estimate noise covarariance
noise_cov = mne.compute_covariance(epochs, tmax=0, method='shrunk')
Out:
ds directory : /home/ubuntu/mne_data/MNE-spm-face/MEG/spm/SPM_CTF_MEG_example_faces1_3D.ds
res4 data read.
hc data read.
Separate EEG position data file not present.
Quaternion matching (desired vs. transformed):
-0.90 72.01 0.00 mm <-> -0.90 72.01 0.00 mm (orig : -43.09 61.46 -252.17 mm) diff = 0.000 mm
0.90 -72.01 0.00 mm <-> 0.90 -72.01 0.00 mm (orig : 53.49 -45.24 -258.02 mm) diff = 0.000 mm
98.30 0.00 0.00 mm <-> 98.30 -0.00 0.00 mm (orig : 78.60 72.16 -241.87 mm) diff = 0.000 mm
Coordinate transformations established.
Polhemus data for 3 HPI coils added
Device coordinate locations for 3 HPI coils added
Measurement info composed.
Finding samples for /home/ubuntu/mne_data/MNE-spm-face/MEG/spm/SPM_CTF_MEG_example_faces1_3D.ds/SPM_CTF_MEG_example_faces1_3D.meg4:
System clock channel is available, checking which samples are valid.
1 x 324474 = 324474 samples from 340 chs
Current compensation grade : 3
Reading 0 ... 324473 = 0.000 ... 675.985 secs...
Setting up band-pass filter from 1 - 30 Hz
172 events found
Events id: [1 2 3]
168 matching events found
0 projection items activated
Loading data for 168 events and 97 original time points ...
Rejecting epoch based on MAG : [u'MLT35-2908', u'MLT42-2908', u'MLT45-2908', u'MLT52-2908', u'MRT14-2908', u'MRT43-2908', u'MRT44-2908', u'MRT45-2908', u'MRT53-2908', u'MRT54-2908']
1 bad epochs dropped
Fitting ICA to data using 274 channels.
Please be patient, this may take some time
Inferring max_pca_components from picks.
Rejecting epoch based on MAG : [u'MLT35-2908', u'MLT42-2908', u'MLT45-2908', u'MLT52-2908', u'MRT14-2908', u'MRT43-2908', u'MRT44-2908', u'MRT45-2908', u'MRT53-2908', u'MRT54-2908']
Artifact detected in [23040, 23280]
Rejecting epoch based on MAG : [u'MLT35-2908', u'MLT42-2908', u'MLT45-2908', u'MLT52-2908', u'MRT14-2908', u'MRT43-2908', u'MRT44-2908', u'MRT45-2908', u'MRT53-2908', u'MRT54-2908']
Artifact detected in [23280, 23520]
Rejecting epoch based on MAG : [u'MRT43-2908']
Artifact detected in [23520, 23760]
Rejecting epoch based on MAG : [u'MLT24-2908', u'MLT25-2908', u'MLT34-2908', u'MLT35-2908']
Artifact detected in [80400, 80640]
Rejecting epoch based on MAG : [u'MLF54-2908', u'MLO11-2908', u'MLT14-2908', u'MLT23-2908', u'MLT24-2908', u'MLT25-2908', u'MLT34-2908', u'MLT35-2908', u'MLT36-2908', u'MRO12-2908', u'MRO32-2908', u'MRO43-2908', u'MRP41-2908']
Artifact detected in [80640, 80880]
Selection by explained variance: 23 components
Using channel MRT31-2908 as EOG channel
EOG channel index for this subject is: [274]
Filtering the data to remove DC offset to help distinguish blinks from saccades
Setting up band-pass filter from 2 - 45 Hz
Setting up band-pass filter from 1 - 10 Hz
Now detecting blinks and generating corresponding events
Number of EOG events detected : 148
148 matching events found
Loading data for 148 events and 121 original time points ...
Rejecting epoch based on MAG : [u'MLT35-2908', u'MLT42-2908', u'MLT45-2908', u'MLT52-2908', u'MRT14-2908', u'MRT43-2908', u'MRT44-2908', u'MRT45-2908', u'MRT53-2908', u'MRT54-2908']
Rejecting epoch based on MAG : [u'MLT35-2908', u'MLT42-2908', u'MLT45-2908', u'MLT52-2908', u'MRT14-2908', u'MRT43-2908', u'MRT44-2908', u'MRT45-2908', u'MRT53-2908', u'MRT54-2908']
Rejecting epoch based on MAG : [u'MLT24-2908', u'MLT35-2908']
Rejecting epoch based on MAG : [u'MLT24-2908', u'MLT34-2908', u'MLT35-2908']
Rejecting epoch based on MAG : [u'MLT24-2908', u'MLT25-2908', u'MLT34-2908', u'MLT35-2908']
Rejecting epoch based on MAG : [u'MLT24-2908', u'MLT25-2908', u'MLT34-2908', u'MLT35-2908']
Rejecting epoch based on MAG : [u'MLT14-2908', u'MLT24-2908', u'MLT25-2908', u'MLT34-2908', u'MLT35-2908']
Rejecting epoch based on MAG : [u'MLT14-2908', u'MLT24-2908', u'MLT25-2908', u'MLT34-2908', u'MLT35-2908', u'MRO12-2908']
Rejecting epoch based on MAG : [u'MLF54-2908', u'MLO11-2908', u'MLT14-2908', u'MLT23-2908', u'MLT24-2908', u'MLT25-2908', u'MLT34-2908', u'MLT35-2908', u'MLT36-2908', u'MRO12-2908', u'MRO32-2908', u'MRO43-2908', u'MRP41-2908']
Rejecting epoch based on MAG : [u'MLF54-2908', u'MLO11-2908', u'MLT14-2908', u'MLT23-2908', u'MLT24-2908', u'MLT25-2908', u'MLT35-2908', u'MLT36-2908', u'MRO12-2908', u'MRO32-2908', u'MRO43-2908', u'MRP41-2908']
Rejecting epoch based on MAG : [u'MLT14-2908', u'MLT24-2908', u'MLT25-2908', u'MLT35-2908']
Rejecting epoch based on MAG : [u'MLT14-2908']
13 bad epochs dropped
Using channel MRT31-2908 as EOG channel
Transforming to ICA space (23 components)
Zeroing out 1 ICA components
Transforming to ICA space (23 components)
Zeroing out 1 ICA components
Estimating covariance using SHRUNK
Done.
Using cross-validation to select the best estimator.
Number of samples used : 4175
[done]
log-likelihood on unseen data (descending order):
shrunk: -1263.881
selecting best estimator: shrunk
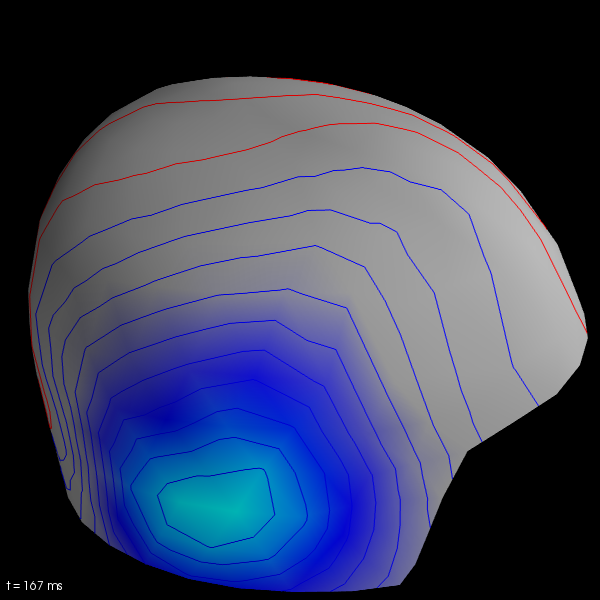
Visualize fields on MEG helmet
trans_fname = data_path + ('/MEG/spm/SPM_CTF_MEG_example_faces1_3D_'
'raw-trans.fif')
maps = mne.make_field_map(evoked[0], trans_fname, subject='spm',
subjects_dir=subjects_dir, n_jobs=1)
evoked[0].plot_field(maps, time=0.170)
Out:
Getting helmet for system CTF_275
Prepare MEG mapping...
Computing dot products for 274 coils...
Computing dot products for 304 surface locations...
Field mapping data ready
Preparing the mapping matrix...
[Truncate at 95 missing 0.0001]
Compute forward model
# Make source space
src_fname = data_path + '/subjects/spm/bem/spm-oct-6-src.fif'
if not op.isfile(src_fname):
src = mne.setup_source_space('spm', src_fname, spacing='oct6',
subjects_dir=subjects_dir, overwrite=True)
else:
src = mne.read_source_spaces(src_fname)
bem = data_path + '/subjects/spm/bem/spm-5120-5120-5120-bem-sol.fif'
forward = mne.make_forward_solution(contrast.info, trans_fname, src, bem)
forward = mne.convert_forward_solution(forward, surf_ori=True)
Out:
Setting up the source space with the following parameters:
SUBJECTS_DIR = /home/ubuntu/mne_data/MNE-spm-face/subjects
Subject = spm
Surface = white
Octahedron subdivision grade 6
Parameters 'fname' and 'overwrite' are deprecated and will be removed in version 0.16. In version 0.15 fname will default to None. Use mne.write_source_spaces instead.
>>> 1. Creating the source space...
Doing the octahedral vertex picking...
Loading /home/ubuntu/mne_data/MNE-spm-face/subjects/spm/surf/lh.white...
Mapping lh spm -> oct (6) ...
Triangle neighbors and vertex normals...
Loading geometry from /home/ubuntu/mne_data/MNE-spm-face/subjects/spm/surf/lh.sphere...
Triangle neighbors and vertex normals...
Setting up the triangulation for the decimated surface...
loaded lh.white 4098/139164 selected to source space (oct = 6)
Loading /home/ubuntu/mne_data/MNE-spm-face/subjects/spm/surf/rh.white...
Mapping rh spm -> oct (6) ...
Triangle neighbors and vertex normals...
Loading geometry from /home/ubuntu/mne_data/MNE-spm-face/subjects/spm/surf/rh.sphere...
Triangle neighbors and vertex normals...
Setting up the triangulation for the decimated surface...
loaded rh.white 4098/137848 selected to source space (oct = 6)
Calculating source space distances (limit=inf mm)...
Computing patch statistics...
Patch information added...
Computing patch statistics...
Patch information added...
Write a source space...
[done]
Write a source space...
[done]
2 source spaces written
Wrote /home/ubuntu/mne_data/MNE-spm-face/subjects/spm/bem/spm-oct-6-src.fif
You are now one step closer to computing the gain matrix
Source space : <SourceSpaces: [<surface (lh), n_vertices=139164, n_used=4098, coordinate_frame=MRI (surface RAS)>, <surface (rh), n_vertices=137848, n_used=4098, coordinate_frame=MRI (surface RAS)>]>
MRI -> head transform source : /home/ubuntu/mne_data/MNE-spm-face/MEG/spm/SPM_CTF_MEG_example_faces1_3D_raw-trans.fif
Measurement data : instance of Info
BEM model : /home/ubuntu/mne_data/MNE-spm-face/subjects/spm/bem/spm-5120-5120-5120-bem-sol.fif
Accurate field computations
Do computations in head coordinates
Free source orientations
Destination for the solution : None
Read 2 source spaces a total of 8196 active source locations
Coordinate transformation: MRI (surface RAS) -> head
0.999622 0.006802 0.026647 -2.80 mm
-0.014131 0.958276 0.285497 6.72 mm
-0.023593 -0.285765 0.958009 9.43 mm
0.000000 0.000000 0.000000 1.00
Read 274 MEG channels from info
Read 29 MEG compensation channels from info
81 coil definitions read
Coordinate transformation: MEG device -> head
0.997940 -0.049681 -0.040594 -1.35 mm
0.054745 0.989330 0.135013 -0.41 mm
0.033453 -0.136957 0.990012 65.80 mm
0.000000 0.000000 0.000000 1.00
5 compensation data sets in info
MEG coil definitions created in head coordinates.
Source spaces are now in head coordinates.
Setting up the BEM model using /home/ubuntu/mne_data/MNE-spm-face/subjects/spm/bem/spm-5120-5120-5120-bem-sol.fif...
Loading surfaces...
Three-layer model surfaces loaded.
Loading the solution matrix...
Loaded linear_collocation BEM solution from /home/ubuntu/mne_data/MNE-spm-face/subjects/spm/bem/spm-5120-5120-5120-bem-sol.fif
Employing the head->MRI coordinate transform with the BEM model.
BEM model spm-5120-5120-5120-bem-sol.fif is now set up
Source spaces are in head coordinates.
Checking that the sources are inside the bounding surface (will take a few...)
Thank you for waiting.
Setting up compensation data...
274 out of 274 channels have the compensation set.
Desired compensation data (3) found.
All compensation channels found.
Preselector created.
Compensation data matrix created.
Postselector created.
Composing the field computation matrix...
Composing the field computation matrix (compensation coils)...
Computing MEG at 8196 source locations (free orientations)...
Finished.
Converting to surface-based source orientations...
Average patch normals will be employed in the rotation to the local surface coordinates....
[done]
Compute inverse solution
snr = 3.0
lambda2 = 1.0 / snr ** 2
method = 'dSPM'
inverse_operator = make_inverse_operator(contrast.info, forward, noise_cov,
loose=0.2, depth=0.8)
# Compute inverse solution on contrast
stc = apply_inverse(contrast, inverse_operator, lambda2, method, pick_ori=None)
# stc.save('spm_%s_dSPM_inverse' % constrast.comment)
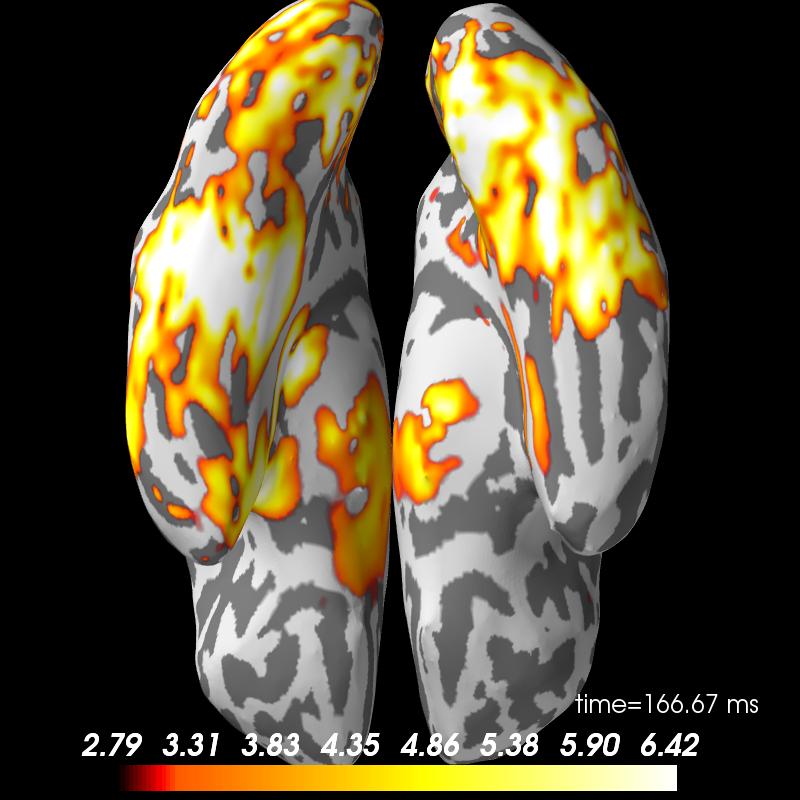
# Plot contrast in 3D with PySurfer if available
brain = stc.plot(hemi='both', subjects_dir=subjects_dir, initial_time=0.170,
views=['ven'])
# brain.save_image('dSPM_map.png')
Out:
Computing inverse operator with 274 channels.
estimated rank (mag): 274
Setting small MEG eigenvalues to zero.
Not doing PCA for MEG.
Total rank is 274
Creating the depth weighting matrix...
274 magnetometer or axial gradiometer channels
limit = 8109/8196 = 10.042069
scale = 4.04483e-11 exp = 0.8
Computing inverse operator with 274 channels.
Creating the source covariance matrix
Applying loose dipole orientations. Loose value of 0.2.
Whitening the forward solution.
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 12.6185
scaling factor to adjust the trace = 3.3072e+19
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 42
Created the regularized inverter
The projection vectors do not apply to these channels.
Created the whitener using a full noise covariance matrix (0 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Picked 274 channels from the data
Computing inverse...
(eigenleads need to be weighted)...
combining the current components...
(dSPM)...
[done]
Updating smoothing matrix, be patient..
Smoothing matrix creation, step 1
Smoothing matrix creation, step 2
Smoothing matrix creation, step 3
Smoothing matrix creation, step 4
Smoothing matrix creation, step 5
Smoothing matrix creation, step 6
Smoothing matrix creation, step 7
Smoothing matrix creation, step 8
Smoothing matrix creation, step 9
Smoothing matrix creation, step 10
colormap: fmin=2.79e+00 fmid=3.17e+00 fmax=6.42e+00 transparent=1
Updating smoothing matrix, be patient..
Smoothing matrix creation, step 1
Smoothing matrix creation, step 2
Smoothing matrix creation, step 3
Smoothing matrix creation, step 4
Smoothing matrix creation, step 5
Smoothing matrix creation, step 6
Smoothing matrix creation, step 7
Smoothing matrix creation, step 8
Smoothing matrix creation, step 9
Smoothing matrix creation, step 10
colormap: fmin=2.79e+00 fmid=3.17e+00 fmax=6.42e+00 transparent=1
Total running time of the script: ( 15 minutes 19.252 seconds)