import numpy as np
import mne
from mne.datasets import sample
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw = mne.io.read_raw_fif(raw_fname)
raw.set_eeg_reference()
Out:
Opening raw data file /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Current compensation grade : 0
An average reference projection was already added. The data has been left untouched.
Sometimes some MEG or EEG channels are not functioning properly for various reasons. These channels should be excluded from analysis by marking them bad as. This is done by setting the ‘bads’ in the measurement info of a data container object (e.g. Raw, Epochs, Evoked). The info[‘bads’] value is a Python string. Here is example:
raw.info['bads'] = ['MEG 2443']
Why setting a channel bad?: If a channel does not show a signal at all (flat) it is important to exclude it from the analysis. If a channel as a noise level significantly higher than the other channels it should be marked as bad. Presence of bad channels can have terribe consequences on down stream analysis. For a flat channel some noise estimate will be unrealistically low and thus the current estimate calculations will give a strong weight to the zero signal on the flat channels and will essentially vanish. Noisy channels can also affect others when signal-space projections or EEG average electrode reference is employed. Noisy bad channels can also adversely affect averaging and noise-covariance matrix estimation by causing unnecessary rejections of epochs.
Recommended ways to identify bad channels are:
mne.io.Raw.plot() without SSP/ICA
enabled and identify bad channels.Note
Setting the bad channels should be done as early as possible in the analysis pipeline. That’s why it’s recommended to set bad channels the raw objects/files. If present in the raw data files, the bad channel selections will be automatically transferred to averaged files, noise-covariance matrices, forward solution files, and inverse operator decompositions.
The actual removal happens using pick_types with
exclude=’bads’ option (see Obtaining subsets of channels).
Instead of removing the bad channels, you can also try to repair them. This is done by interpolation of the data from other channels. To illustrate how to use channel interpolation let us load some data.
# Reading data with a bad channel marked as bad:
fname = data_path + '/MEG/sample/sample_audvis-ave.fif'
evoked = mne.read_evokeds(fname, condition='Left Auditory',
baseline=(None, 0))
# restrict the evoked to EEG and MEG channels
evoked.pick_types(meg=True, eeg=True, exclude=[])
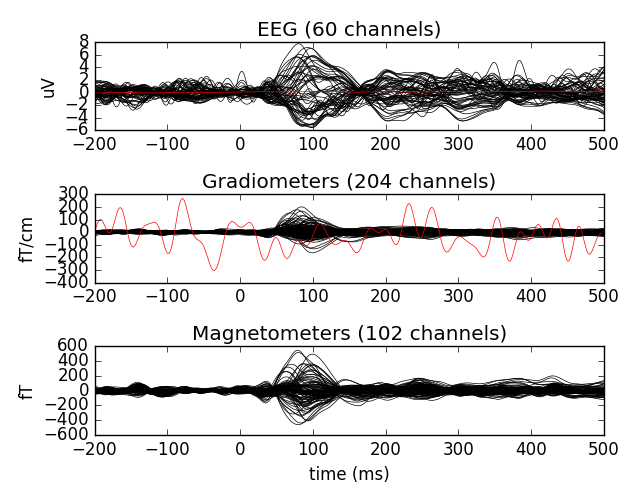
# plot with bads
evoked.plot(exclude=[])
print(evoked.info['bads'])
Out:
Reading /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
[u'MEG 2443', u'EEG 053']
Let’s now interpolate the bad channels (displayed in red above)
evoked.interpolate_bads(reset_bads=False)
Out:
Computing interpolation matrix from 59 sensor positions
Interpolating 1 sensors
Computing dot products for 305 coils...
Computing cross products for coils 305 x 1 coils...
Preparing the mapping matrix...
[Truncate at 79 missing 0.0001]
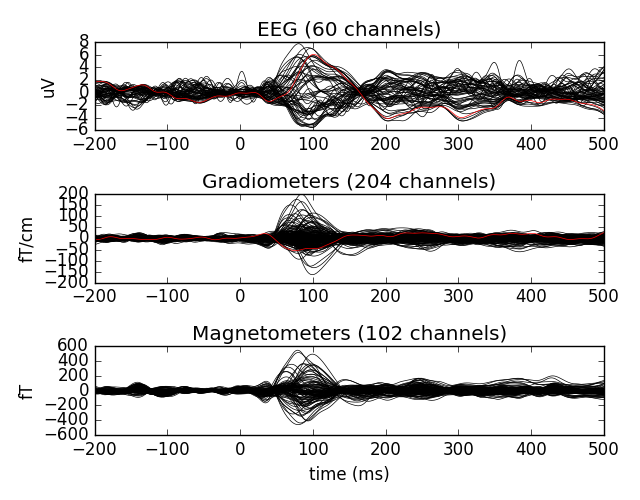
Let’s plot the cleaned data
evoked.plot(exclude=[])
Note
Interpolation is a linear operation that can be performed also on Raw and Epochs objects.
For more details on interpolation see the page Repairing bad channels.
MNE provides an mne.Annotations class that can be used to mark
segments of raw data and to reject epochs that overlap with bad segments
of data. The annotations are automatically synchronized with raw data as
long as the timestamps of raw data and annotations are in sync.
See Brainstorm auditory tutorial dataset for a long example exploiting the annotations for artifact removal.
The instances of annotations are created by providing a list of onsets and
offsets with descriptions for each segment. The onsets and offsets are marked
as seconds. onset refers to time from start of the data. offset is
the duration of the annotation. The instance of mne.Annotations
can be added as an attribute of mne.io.Raw.
eog_events = mne.preprocessing.find_eog_events(raw)
n_blinks = len(eog_events)
# Center to cover the whole blink with full duration of 0.5s:
onset = eog_events[:, 0] / raw.info['sfreq'] - 0.25
duration = np.repeat(0.5, n_blinks)
raw.annotations = mne.Annotations(onset, duration, ['bad blink'] * n_blinks,
orig_time=raw.info['meas_date'])
raw.plot(events=eog_events) # To see the annotated segments.
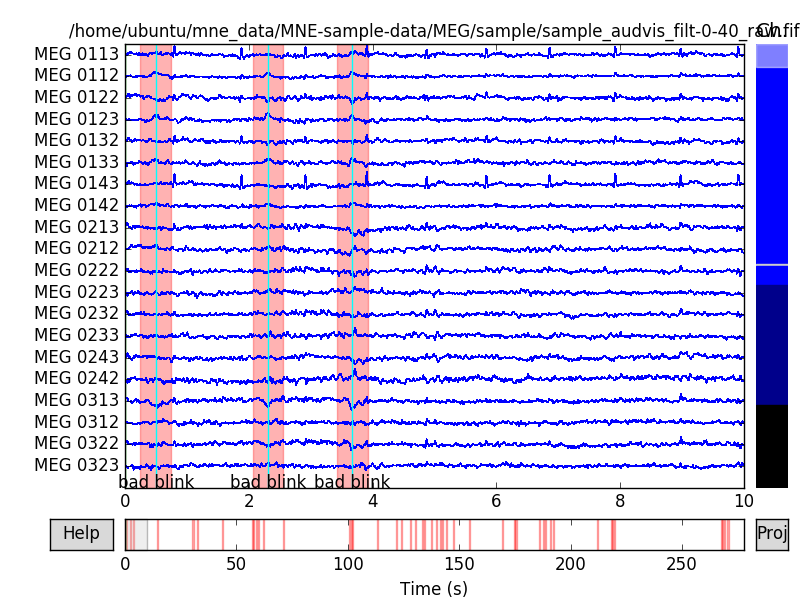
Out:
EOG channel index for this subject is: [375]
Filtering the data to remove DC offset to help distinguish blinks from saccades
Setting up band-pass filter from 2 - 45 Hz
Setting up band-pass filter from 1 - 10 Hz
Now detecting blinks and generating corresponding events
Number of EOG events detected : 46
It is also possible to draw bad segments interactively using
raw.plot (see Visualize Raw data).
As the data is epoched, all the epochs overlapping with segments whose
description starts with ‘bad’ are rejected by default. To turn rejection off,
use keyword argument reject_by_annotation=False when constructing
mne.Epochs. When working with neuromag data, the first_samp
offset of raw acquisition is also taken into account the same way as with
event lists. For more see mne.Epochs and mne.Annotations.
When working with segmented data (Epochs) MNE offers a quite simple approach to automatically reject/ignore bad epochs. This is done by defining thresholds for peak-to-peak amplitude and flat signal detection.
In the following code we build Epochs from Raw object. One of the provided parameter is named reject. It is a dictionary where every key is a channel type as a sring and the corresponding values are peak-to-peak rejection parameters (amplitude ranges as floats). Below we define the peak-to-peak rejection values for gradiometers, magnetometers and EOG:
reject = dict(grad=4000e-13, mag=4e-12, eog=150e-6)
Note
The rejection values can be highly data dependent. You should be careful when adjusting these values. Make sure not too many epochs are rejected and look into the cause of the rejections. Maybe it’s just a matter of marking a single channel as bad and you’ll be able to save a lot of data.
We then construct the epochs
events = mne.find_events(raw, stim_channel='STI 014')
event_id = {"auditory/left": 1}
tmin = -0.2 # start of each epoch (200ms before the trigger)
tmax = 0.5 # end of each epoch (500ms after the trigger)
baseline = (None, 0) # means from the first instant to t = 0
picks_meg = mne.pick_types(raw.info, meg=True, eeg=False, eog=True,
stim=False, exclude='bads')
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, proj=True,
picks=picks_meg, baseline=baseline, reject=reject,
reject_by_annotation=True)
Out:
319 events found
Events id: [ 1 2 3 4 5 32]
72 matching events found
Created an SSP operator (subspace dimension = 3)
4 projection items activated
We then drop/reject the bad epochs
epochs.drop_bad()
Out:
Loading data for 72 events and 106 original time points ...
Rejecting epoch based on EOG : [u'EOG 061']
Rejecting epoch based on MAG : [u'MEG 1711']
22 bad epochs dropped
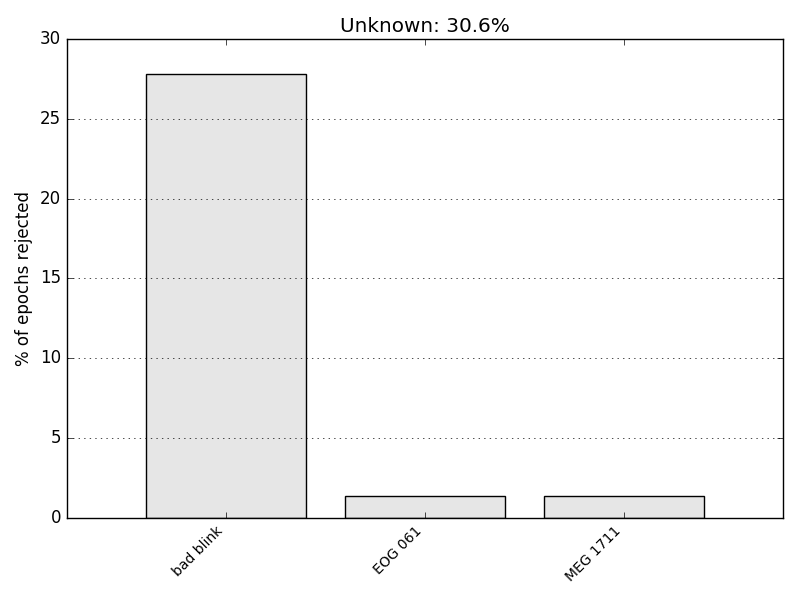
And plot the so-called drop log that details the reason for which some epochs have been dropped.
print(epochs.drop_log[40:45]) # only a subset
epochs.plot_drop_log()
Out:
[['bad blink'], ['IGNORED'], ['IGNORED'], ['IGNORED'], ['bad blink']]
What you see is that some drop log values are empty. It means event was kept. If it says ‘IGNORED’ is means the event_id did not contain the associated event. If it gives the name of channel such as ‘EOG 061’ it means the epoch was rejected because ‘EOG 061’ exceeded the peak-to-peak rejection limit.
Total running time of the script: ( 0 minutes 11.227 seconds)