Contents
This Chapter gives a detailed description of the processing of a sample data set, which can be employed to familiarize with the workflow described in Cookbook.
Note
Going through the analysis exercise in this chapter is not a substitute for reading other chapters of this manual and understanding the concepts underlying MNE software.
The MNE software is accompanied by a sample data set which includes the MRI reconstructions created with FreeSurfer and the an MEG/EEG data set. These data were acquired with the Neuromag Vectorview system at MGH/HMS/MIT Athinoula A. Martinos Center Biomedical Imaging. EEG data from a 60-channel electrode cap was acquired simultaneously with the MEG. The original MRI data set was acquired with a Siemens 1.5 T Sonata scanner using an MPRAGE sequence.
Note
These data are provided solely for the purpose of getting familiar with the MNE software. They should not be redistributed to third parties. The data should not be used to evaluate the performance of the MEG or MRI system employed.
In the MEG/EEG experiment, checkerboard patterns were presented into the left and right visual field, interspersed by tones to the left or right ear. The interval between the stimuli was 750 ms. Occasionally a smiley face was presented at the center of the visual field. The subject was asked to press a key with the right index finger as soon as possible after the appearance of the face. A listing of the corresponding trigger codes is provided in Trigger codes for the sample data set.
| Name | #Contents | |
|---|---|---|
| LA | 1 | Response to left-ear auditory stimulus |
| RA | 2 | Response to right-ear auditory stimulus |
| LV | 3 | Response to left visual field stimulus |
| RV | 4 | Response to right visual field stimulus |
| smiley | 5 | Response to the smiley face |
| button | 32 | Response triggered by the button press |
The sample dataset can be downloaded automatically by doing:
>>> mne.datasets.sample.data_path(verbose=True)
The sample data set contains two main directories: MEG/sample (the MEG/EEG
data) and subjects/sample (the MRI reconstructions).
An overview of the data provided is given in Tables Contents of the MEG/sample directory. and Overview of the contents of the subjects/sample directory.. In addition to
subject sample , the MRI surface
reconstructions from another subject, morph ,
are provided to demonstrate the morphing capabilities of the MNE software.
| File | Contents |
|---|---|
| sample/audvis_raw.fif | The raw MEG/EEG data |
| audvis.ave | A template script for off-line averaging |
| auvis.cov | A template script for the computation of a noise-covariance matrix |
| File / directory | Contents |
|---|---|
| bem | Directory for the forward modelling data |
| bem/watershed | BEM surface segmentation data computed with the watershed algorithm |
| bem/inner_skull.surf | Inner skull surface for BEM |
| bem/outer_skull.surf | Outer skull surface for BEM |
| bem/outer_skin.surf | Skin surface for BEM |
| sample-head.fif | Skin surface in fif format for mne_analyze visualizations |
| surf | Surface reconstructions |
| mri/T1 | The T1-weighted MRI data employed in visualizations |
The following preprocessing steps have been already accomplished in the sample data set:
To set up the structural MRIs for processing with the Neuromag MRI viewer, MRIlab, say
mne_setup_mri
This command sets up the directories subjects/sample/mri/T1-neuromag and subjects/sample/mri/brain-neuromag .
For more information, see Setting up anatomical MR images for MRIlab.
The source space with a 5-mm grid spacing is set up by saying:
mne_setup_source_space --ico -6
This command sets up the source-space related files in directory subjects/sample/bem as
described in Setting up the source space.
The geometry calculations for the single-layer boundary-element model are accomplished with the command:
mne_setup_forward_model --homog --surf --ico 4
This command sets up the homogeneous BEM-model related files
in directory subjects/sample/bem as described in Setting up the boundary-element model. In
addition to the homogeneous BEM, you also need the three-layer BEM model,
which can be used for both EEG and MEG:
mne_setup_forward_model --surf --ico 4
The above commands employ the inner_skull.surf , outer_skull.surf ,
and outer_skin.surf triangulation files located in subjects/sample/bem .
The option --ico 4 will create a model with 5120 triangles
on each surface. Depending on the speed of your computer, the three-layer
model may take quite a while to set up.
A data specific EEG layout will facilitate viewing of the
EEG data. The MNE programs mne_browse_raw and mne_analyze look
for user-specific layouts in $HOME/.mne/lout . Thus,
you can create an EEG layout for the sample data set with the following
commands:
mkdir -p $HOME/.mne/lout
cd $SAMPLE/MEG/sample
mne_make_eeg_layout --fif sample_audvis_raw.fif --lout $HOME/.mne/lout/sample-EEG.lout
Please refer to mne_make_eeg_layout for more information on mne_make_eeg_layout .
Note
It is usually sufficient to create one EEG layout for each electrode cap you are using in your experiment rather than using a different layout file for each data file generated using the same cap.
Previewing your raw data before proceeding to averaging and computation of the current estimates is most important to avoid unintentional errors caused by noisy or dysfunctional channels, frequent eye blinks, inappropriate bandpass filtering etc.
One possible routemap for the preview session is outlined below:
cd $SAMPLE/MEG/sample .sample_audvis_raw.fif from File/Open… .After these steps, you are ready to proceed to the actual analysis.
Go to directory $SAMPLE/MEG/sample . With help
of Description files for off-line averaging, familiarize yourself with the averaging
script audvis.ave .
You can invoke an averaging script in mne_browse_raw from Process/Average… .
Select the audvis.ave script from the file selection
box that appears. Once averaging is complete, you can inspect the
details of the averaged responses in the Averages window,
which appears automatically. You can redisplay it from Windows/Show averages… . The window, which appears when you
select Adjust/Manage averages… allows
you to:
Note
If you decide to save the averages in the interactive mode, use the name sample_audvis-ave.fif for the result.
The batch-mode version of mne_browse_raw , mne_process_raw can be used for averaging as well. Batch mode averaging can be done with the command:
mne_process_raw --raw sample_audvis_raw.fif `` ``--lowpass 40 --projoff `` ``--saveavetag -ave --ave audvis.ave
See mne_process_raw for command-line options.
As a result of running the averaging script a file called sample_audvis-ave.fif is
created. It contains averages to the left and right ear auditory
as well as to the left and right visual field stimuli.
The average file computed in the previous section can be viewed in mne_browse_raw .
To view the averaged signals, invoke mne_browse_raw :
cd $SAMPLE/MEG/sample
mne_browse_raw &
This Section gives only very basic information about the use of mne_browse_raw for viewing evoked-response data. Please consult Browsing raw data with mne_browse_raw for more comprehensive information.
mne_browse_raw loads all the available data from an average file at once:
sample_audvis-ave.fif file
from the list and click OK .Select the left and right ear auditory stimulus responses for display:
Set the time scale and baseline:
You can display a subset of responses from the topographical display by holding the shift key down and dragging with the mouse, left button down. When you drag on the response with just the left button down, the signal timing, and channel name are displayed at the bottom. If the left mouse button is down and you press shift down the time is give both in absolute units and relative to the point where shift was pressed down.
Observe the following:
Go back to the Manage averages… dialog and switch all other conditions except the visual ones.
Observe the following:
Note
If you have the Neuromag software available, the averaged data can be also viewed in the Neuromag data plotter (xplotter ). See Using Neuromag software for instructions on how to use the Neuromag software at the MGH Martinos Center.
Another piece of information derived from the raw data file is the estimate for the noise-covariance matrix, which can be computed with the command:
mne_process_raw --raw sample_audvis_raw.fif --lowpass 40 --projon --savecovtag -cov --cov audvis.cov
Using the definitions in audvis.cov , this command
will create the noise-covariance matrix file sample_audvis-cov.fif .
In this case the projections are set on. The projection information
is then attached to the noise-covariance matrix and will be automatically
loaded when the inverse-operator decomposition is computed.
Note
You can study the contents of the covariance matrix computation description file audvis.cov with the help of Description files for covariance matrix estimation.
The mne_analyze module of the MNE is one option for the coordinate alignment. It uses a triangulated scalp surface to facilitate the alignment.
Follow these steps to make an initial approximation for the coordinate alignment.
MEG/sample .sample_audvis_raw.fif .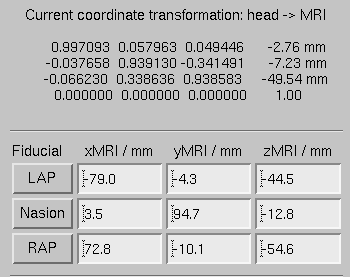
Example of an initial coordinate alignment.
Before proceeding to the refinement procedure, it is useful to remove outlier digitizer points. When you rotate the image in the viewer window, you will notice that there is at least one such point over the right cheek. To discard this point:
The coordinate transformation can be adjusted manually with the arrow buttons in the middle part of the Adjust coordinate alignment dialog. These buttons move the digitizer points in the directions indicated by the amount listed next to each of the buttons.
An automatic iterative procedure, Iterative Closest Point (ICP) matching is also provided. At each iteration step
In step 2 of the iteration, the nasion is assigned five times the weight of the other points since it can be assumed that the nasion is the easiest point to identify reliably from the surface image.
The ICP alignment can be invoked by entering the desired number of iterations next to the ICP align button followed by return or simply pressing the ICP align button. The iteration will converge in 10 to 20 steps.
Warning
Use the ICP alignment option in mne_analyze with caution. The iteration will not converge to a reasonable solution unless and initial alignment is performed first according to Initial alignment. Outlier points should be excluded as described above. No attempt is made to compensate for the possible distance of the digitized EEG electrode locations from the scalp.
To create a MRI fif description file which incorporates the
coordinate transformation click Save MRI set in
the Adjust coordinate alignment dialog.
This will create the MRI set file in the $SUBJECTS_DIR/sample/mri/T1-neuromag/sets directory,
which was created by mne_setup_mri_data ,
see Structural MRIs. The file will be called
COR- <username>- <date>- <time> .fif
where <username> is your login name.
You can also save transformation to a fif file through the Save… button. If the file does not exist, it will only contain the coordinate transformation. If the file exists it will be inserted to the appropriate context. An existing transformation will not be replaced unless Overwrite existing transform is checked in the save dialog.
Once you have saved the coordinate transformation, press Done and quit mne_analyze (File/Quit ).
Note
If you dismiss the alignment dialog before saving the transformation, it will be lost.
To compute the forward solution, say:
cd $SAMPLE/MEG/sample
mne_do_forward_solution --mindist 5 --spacing oct-6 --bem sample-5120-5120-5120 --meas sample_audvis-ave.fif
This produces an EEG and MEG forward solution with source
space points closer than 5 mm to the inner skull surface omitted.
The source space created in Source space will be employed.
As the output from this command will indicate The forward solution
will be stored in file sample_audvis-ave-oct-6-fwd.fif .
This command uses the three-layer BEM model sample-5120-5120-5120-bem-sol.fif created
in Boundary-element models. If you want to use the single-compartment
BEM sample-5120-bem-sol.fif usable for MEG data only
say:
cd $SAMPLE/MEG/sample
mne_do_forward_solution --mindist 5 --spacing oct-6 --meas sample_audvis-ave.fif --bem sample-5120 --megonly
The inverse operator information, necessary for the computation of the MNEs and dSPMs is accomplished by the command:
mne_do_inverse_operator --fwd sample_audvis-ave-oct-6-fwd.fif --depth --loose 0.2 --meg --eeg
This produces a depth-weighted inverse operator decomposition with ‘loose’ orientation constraint applied. More details on the convenience script mne_do_inverse_operator are provided in Calculating the inverse operator.
The above command employs both EEG and MEG data. To create separate solution for EEG and MEG, run the commands:
mne_do_inverse_operator --fwd sample_audvis-ave-oct-6-fwd.fif --depth --loose 0.2 --meg
and
mne_do_inverse_operator --fwd sample_audvis-ave-oct-6-fwd.fif --depth --loose 0.2 --eeg
Note
If you were using a single-compartment BEM to compute the forward solution, you can only compute the MEG inverse operator.
The most exciting part of this exercise is to explore the data and the current estimates in mne_analyze . This section contains some useful steps to get you started. A lot of information about the capabilities of mne_analyze is given in Interactive analysis with mne_analyze. Batch-mode processing with mne_make_movie is discussed in Producing movies and snapshots. Cross-subject averaging is covered in Morphing and averaging.
Before launching mne_analyze it
is advisable to go to the directory MEG/sample . The
current working directory can be also changed from mne_analyze .
Launch mne_analyze . Select Help/On GLX… , which brings up a window containing Open GL rendering context information. If first line in the information dialog that pops up says Nondirect rendering context instead of Direct rendering context you will experience slow graphics performance. To fix this, your system software, graphics adapter or both need to be updated. Consult a computer support person for further information.
It is reasonable to start the analysis by loading the display surfaces: choose the inflated surface for subject sample from the dialog that appears when you select File/Load surface… .
Select File/Open… . Select sample_audvis-ave.fif as
your data file and select the Left auditory data
set. Select the inverse operator sample_audvis-ave-oct-6-meg-eeg-inv.fif and
press OK . After a while the signals
appear in the sample waveform and topographical displays. Click
on the N100m peak in the auditory response. A dSPM map appears in
the main surface display.
Select Windows/Show viewer… . After a while the viewer window appears. Click on the N100m peak again. Once the field map preparation computations are complete, the magnetic field and potential maps appear. Investigate the viewer window options with help of The viewer.
The options affecting the current estimates are accessible from Adjust/Estimate parameters… . With help of Working with current estimates, investigate the effects of the parameter settings.
While in directory MEG/sample , create a directory
called label :
mkdir label
Using the information in Creating new label files, create two
labels A-lh.label and A-rh.label in the
approximate location of the left and right auditory cortices. Save
these labels in the newly created label directory.
Load all labels from the label directory and
investigate the timecourses in these two labels as well as at individual
vertices. Information on label processing can be found from Inquiring timecourses.
Goto to $SUBJECTS_DIR and create the directory morph-maps .
Load the inflated surface for subject morph as
the morphing surfaces. Try switching between the original and morphing
surfaces. More information about morphing is available in Morphing and
in Morphing and averaging.
There is also a left-hemisphere occipital patch file available for subject morph . Load a righ-hemifield visual response instead of the auditory one and investigate mapping of the current estimates on the patch.